Metadata is standardized information that describes and organizes data, essentially “data about data.” It ensures data can be understood, discovered, accessed, and reused efficiently.
Purpose of Metadata
-
Understandability: Adds context, such as study name, assay type, or species, making data comprehensible even to those not involved in its generation.
-
Discovery: Standardized metadata makes data searchable and accessible through organized systems.
-
Interoperability: Ensures data can integrate with other applications smoothly.
-
Reusability: Enables effective data reuse by improving understanding, discovery, and compatibility.
Metadata Structure in the ELITE Portal
Metadata in the ELITE Portal is curated into three main files:
-
Individual Metadata: Contains participant details (e.g., sex, diagnosis) linked by individualID.
-
Biospecimen Metadata: Details specimens collected (e.g., organ, tissue type) linked by specimenID.
-
Assay Metadata: Describes methods used on specimens (e.g., platform, quality control).
These files can be joined using individualID and specimenID for comprehensive insights.
A subset of metadata is stored as annotations on data files, while the complete metadata is saved in separate .CSV files. These files provide detailed information about individuals, specimens (biosamples), and the assays performed.
What is in each metadata file?
-
Individual Metadata
Contains details about study participants, such as sex and diagnosis. Each row represents a unique participant identified by the key individualID.
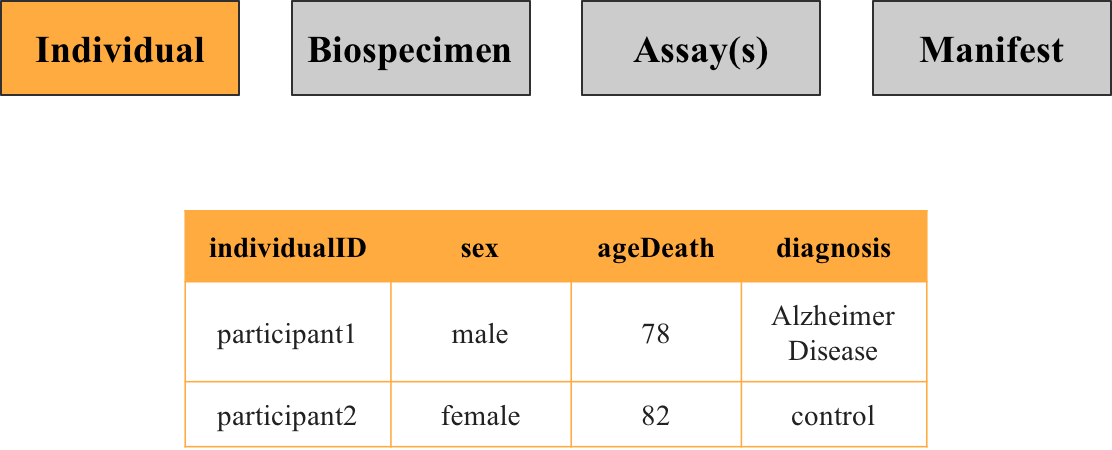
2. Biospecimen metadata
Describes specimens collected from participants, including the source organ or tissue. Each row represents a unique specimen identified by the key specimenID, with individuals potentially linked to multiple specimens.
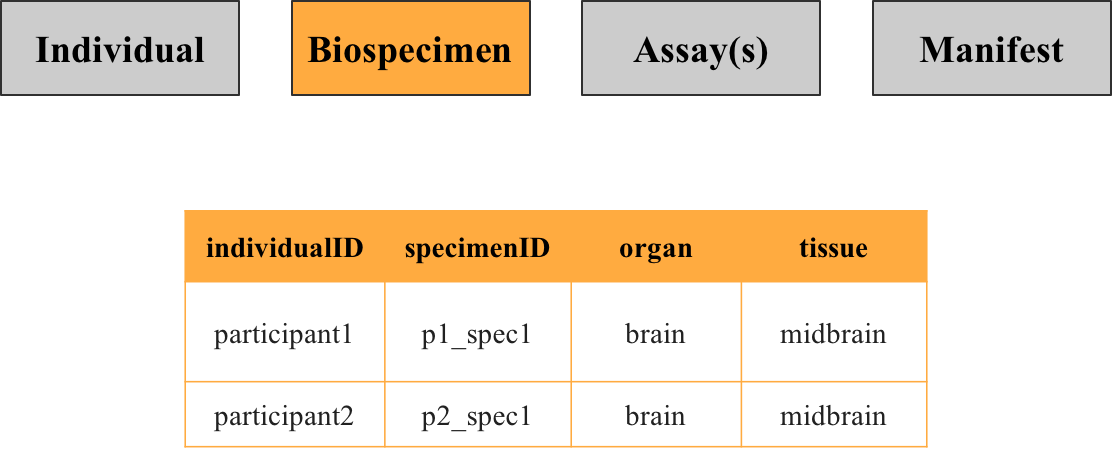
3. Assay(s) metadata
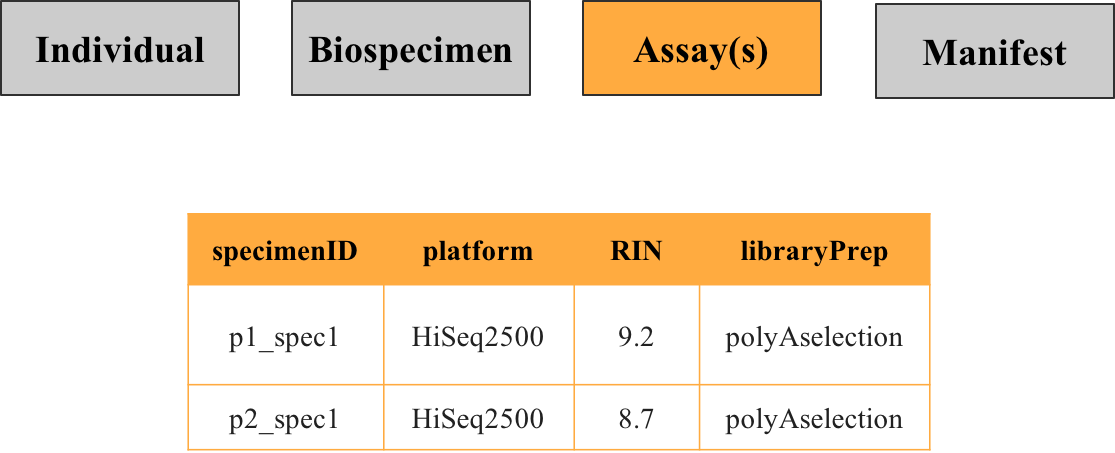
Provides details about procedures performed on specimens to generate data, such as the technology used, batch processing, or quality control metrics. For instance, RNA sequencing metadata would specify the platform used and specimen preparation details. Multiple assay metadata files may exist if specimens or individuals underwent different assay types.
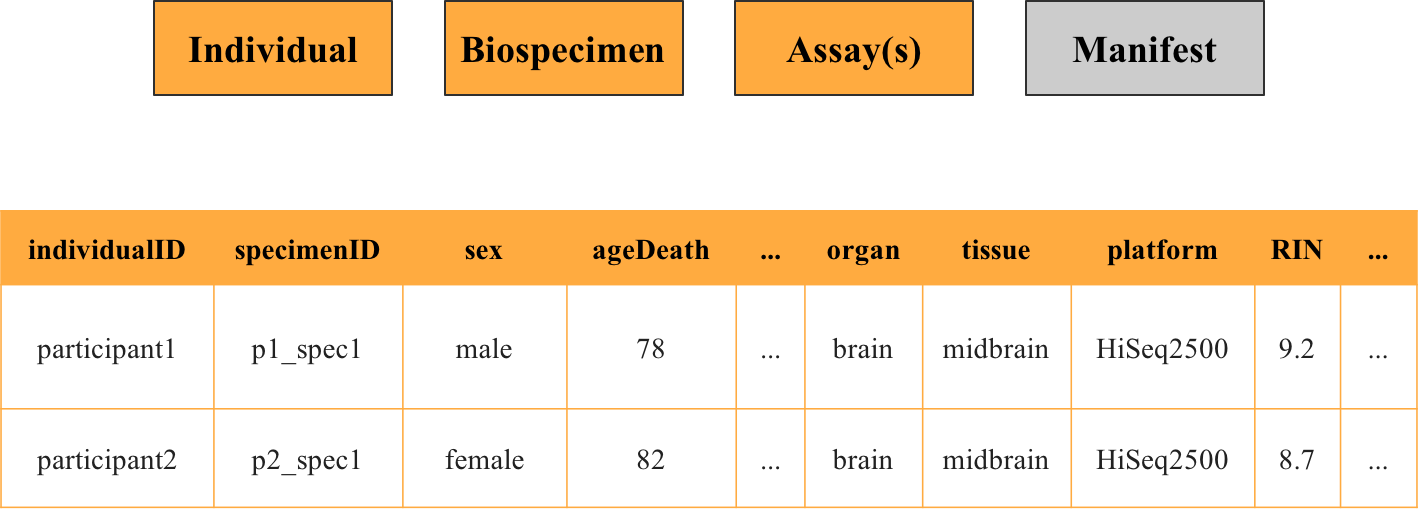
Joining Metadata Files
Metadata files for individuals, biospecimens, and assays can be linked using individualID and specimenID. These connections enable you to combine metadata with the data itself, creating a comprehensive view of each file.
For more on locating and using metadata within the portal, refer to User Guide #1: Find and Download Data, Metadata, and Annotations.
You can also explore User Guide #3: Joining Metadata with Data for a step-by-step guide.
Metadata Dictionary: Overview and Usage
The ELITE Portal Metadata Dictionary is a resource that helps users and contributors understand and use metadata effectively. It provides standardized terms for querying or contributing data, ensuring consistency and clarity.
Who Benefits from the Metadata Dictionary?
-
Users: Helps you understand metadata terms in annotations and files, making it easier to search for and retrieve data.
-
Contributors: Provides accepted values for metadata keys. For example, a HiSeq 2000 machine should be labeled as “HiSeq2000” (no space) under the key platform.
-
Curators: Guides contributors on using correct keys/values and identifies gaps where new terms may be needed.
Using the Metadata Dictionary
The dictionary includes helpful search tools:
-
A global search bar to look up any term.
-
Dedicated column search bars for narrowing results within specific columns.
Scroll to the bottom to navigate between pages if you don’t see all rows at once.
Important Definitions
-
Key: Name of the metadata term.
-
Key Description: Explanation of the term’s purpose.
-
Type: Expected data type (e.g., string, integer).
-
Value: Specific allowed values or a blank field if any text is accepted.
-
Value Description: Details about the value, if applicable.
-
Source: Origin of the value description (e.g., a link or document name).
-
Module: The group or context the term is associated with.